ProGenTomics has over 4 years of expertise with the analysis of ancient remains. In those years, significant contributions have been made to palaeoproteomic field such as the development of a species classification pipeline based on untargeted LC-MS/MS analysis. Our expertise comprises of species classification on bone remains—both on single and mixed bone samples such as coprolites and dust from bone bags, sex determination of human and animal remains, shotgun protein analysis of human remains and protein analysis of pottery food crusts.
Sex determination
 Determining sex of human remains is one of the most crucial factors inreconstructing the past of a given society. The two most common approaches are visual determination and DNA-analysis based on sexual dimorphism of the remains or the sex chromosomes. However, these require intact, non-juvenile remains or the presence of DNA, which is often not the case, especially when dealing with ancient remains. Recently, protein analysis of tooth enamel has entered the scene, enabling sex determination based on the presence of amelogenin Y in male individuals. However, the biggest analytical challenge still remains: proving the absence of AMELY protein in female individuals. We have developed a controlled assay with prior digestion using trypsin and the addition of isotopically labelled internal peptide standards specific for AMELY and AMELX. These standards enable the absolute quantification of both AMELY and AMELX, thus controlling sampling quality. In addition, the presence of isotopically labelled standards also provides a reference signal in the raw data that can recover unidentified AMELY signal, e.g. in the presence of collagen masking the signal, which otherwise impairs sex determination.
Determining sex of human remains is one of the most crucial factors inreconstructing the past of a given society. The two most common approaches are visual determination and DNA-analysis based on sexual dimorphism of the remains or the sex chromosomes. However, these require intact, non-juvenile remains or the presence of DNA, which is often not the case, especially when dealing with ancient remains. Recently, protein analysis of tooth enamel has entered the scene, enabling sex determination based on the presence of amelogenin Y in male individuals. However, the biggest analytical challenge still remains: proving the absence of AMELY protein in female individuals. We have developed a controlled assay with prior digestion using trypsin and the addition of isotopically labelled internal peptide standards specific for AMELY and AMELX. These standards enable the absolute quantification of both AMELY and AMELX, thus controlling sampling quality. In addition, the presence of isotopically labelled standards also provides a reference signal in the raw data that can recover unidentified AMELY signal, e.g. in the presence of collagen masking the signal, which otherwise impairs sex determination.
ProGenTomics is currently developing a new method based on high-throughput peptide enrichment, providing up to a 1000-fold increase in sensitivity compared to the current available workflows. This will limit sample destruction by using a shortened acid etch protocol, increase confidence of the determined sex, and increase sample throughput. Coming soon.
Species classification
Bone morphology-based species classification has been the state-of-the-art for both palaeontological and zooarchaeological research for decades. However, when specimens are degraded, fragmented, and/or fractured to a point where this methodology can no longer be used, paleoproteomic analysis has proven to be invaluable to tackle this challenge. The minute amount of sample required, the automatable sample preparation, the unbiased analysis, and the low analysis cost make it a formidable approach. However, species classification of ancient remains is usually done through ZooMS, which relies on peptide mass fingerprinting based on MALDI-TOF-MS data. Herein species are classified based on the presence of certain collagen peptide markers. This requires a certain amount of prior information, is prone to mistakes, highly fragmented remains are difficult to analyze and peptide marker databases are limited.
LC-MS/MS analysis is increasingly applied for palaeoproteomic species classification. However, these approaches currently rely on curated spectral databases of commonly identified peptides and proteins in archaeological mammalian bones to tackle the complexity of the data (analysis).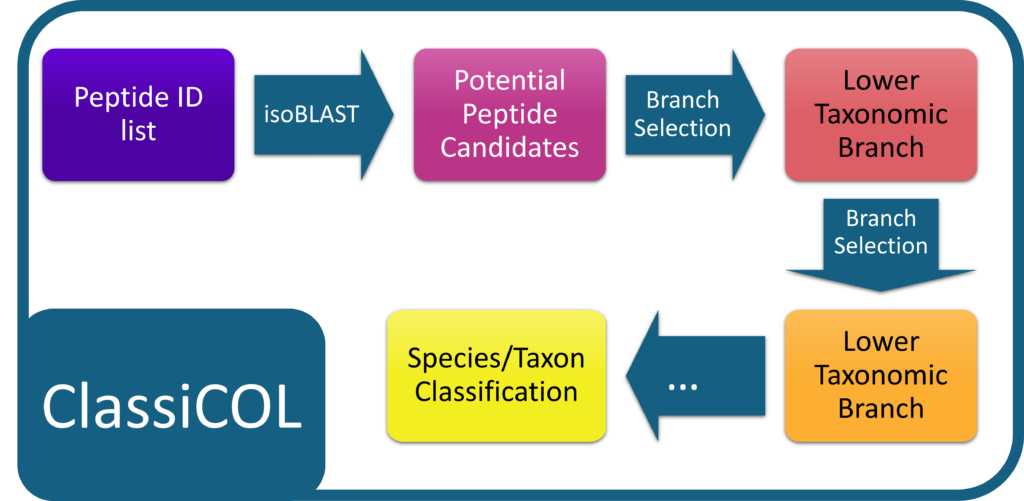
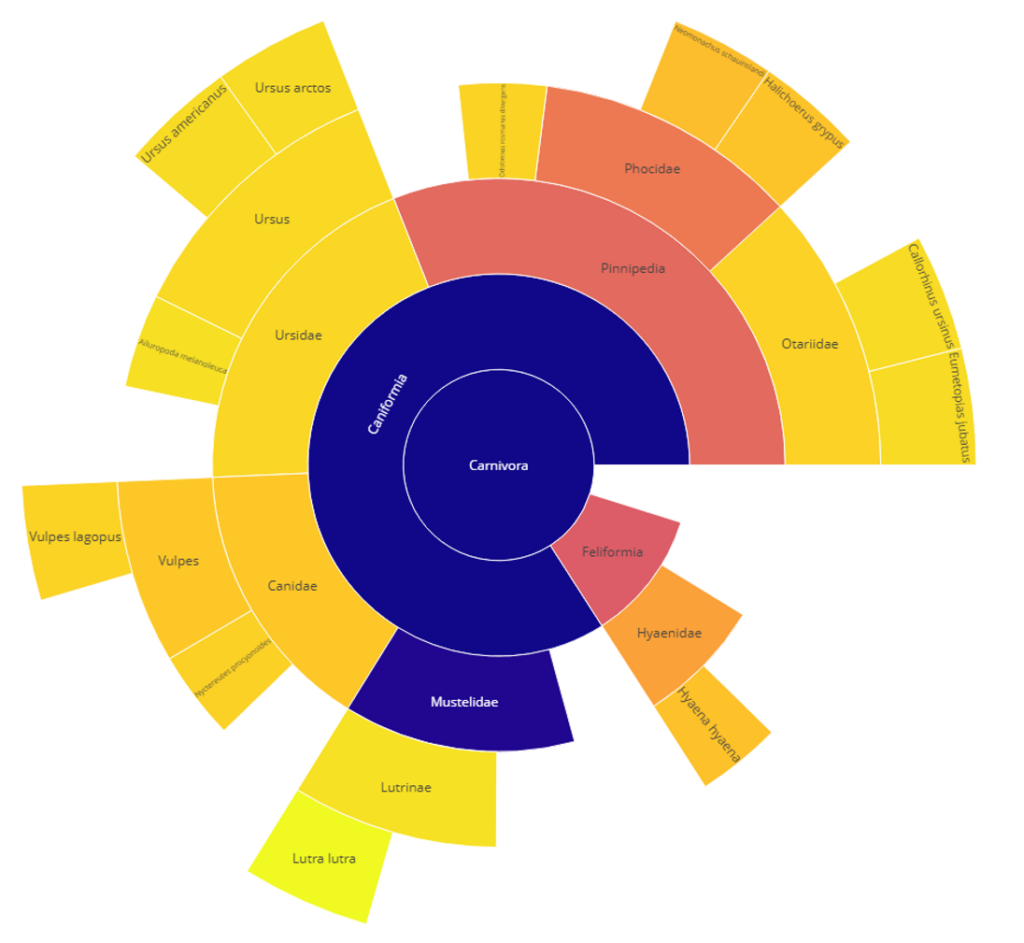
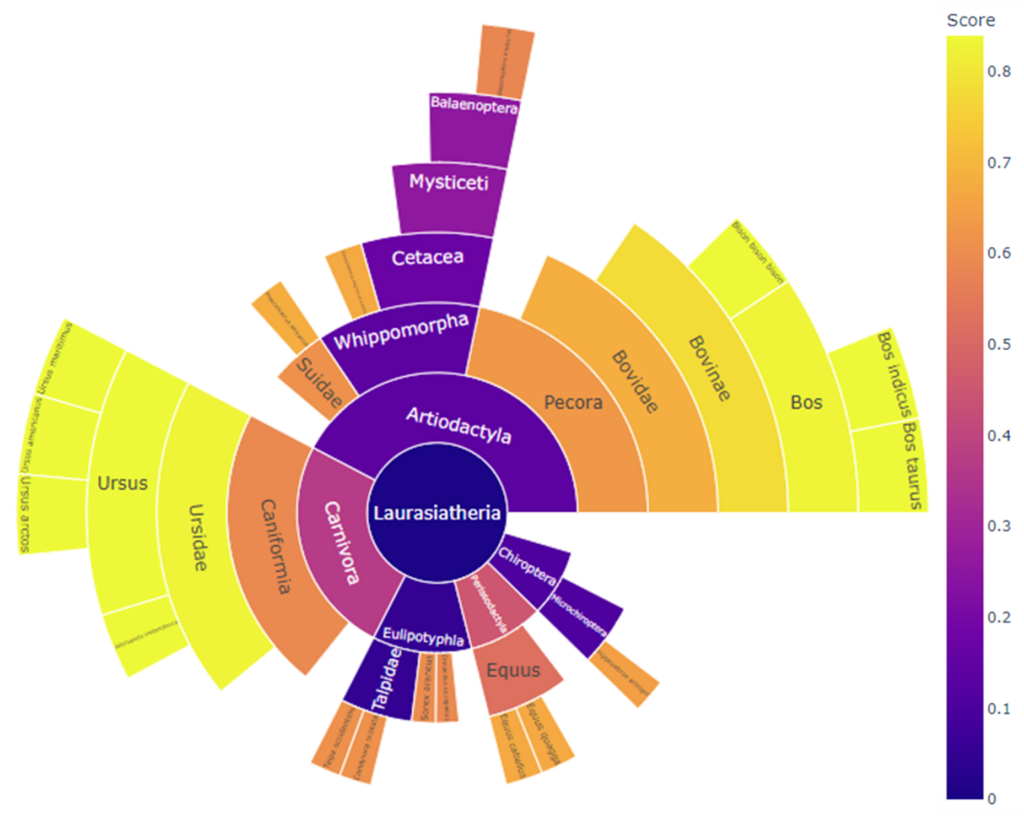
Our ClassiCOL workflow is also based on LC-MS/MS data, but embraces the complexity of the data and extends any peptide identification file to an exhaustive list of potential peptide candidates based on isobaric blasting of the identified peptides. This is done by allowing for local, isobaric amino acid changes and modifications that do not change the precursor mass of the initial peptide. This ensures that the correct answer for any given MS/MS spectrum is in the list of potential peptide candidates. This list is used to run down the NCBI taxonomic tree, retaining one or both branches at each split based on peptide uniqueness – a subset branch is rejected, two different branches are retained, and two identical branches stop the process. The complete list of considered peptides and their respective taxonomic branches are graphically represented in a sunburst plot and the obtained species classifications are scored, giving one or multiple potential species for a given sample. The ClassiCOL workflow not only allows to classify and/or identify species without prior knowledge, but also allows for the identification of mixtures such as coprolites and dust from mixed bone fragment containers.
Example sunburst plot of a single species re-classification into Lutra lutra (otter) after morphological classification as Vulpes vulpes (fox).