Background
Histones are small proteins that contain an above average number of lysine and arginine residues, which makes them overall basic and positively charged. This induces electrostatic interactions with the negatively charged phosphate groups of nuclear DNA, which enable the formation of nucleosomes, the basic unit of chromatin. In 1973, the discovery that chromatin consists of repeating units – later coined nucleosomes for the first time in 1975 – lead to a shift in the conception of chromatin structure. Today it is known that nucleosomes consist of about 147 base pairs of DNA wrapped around an octamer core that contains two copies of each of the four core histones; H2A, H2B, H3 and H4. These nucleosomes are separated by 20-80 base pairs of linker DNA and fixed by the linker histone H1 into the so-called beads-on-a-string formation. This configuration is further condensed into 30nm chromatin fibers and eventually into chromosomes. Through this configuration, histones enable the condensation of the human cellular genome from a 2m stretch of DNA into a 28cm string of nucleosomes, over into chromatin fibers of roughly 180mm and eventually into chromosomes of a few µm in length, which perfectly fit the cell nucleus. However, the classical representation of chromosomes as densely packed rods connected through a centromere only occurs during cell division to ensure the equal distribution of the genome over two daughter cells. After division and for the bigger part of a cell’s life, parts of the chromosome uncoil into a lose state, which enables gene regulation and expression.
For years it was assumed that histones were merely static carriers that facilitate condensation, handling, and manipulation of the vast amounts of DNA in Eukaryotes. However, most cells of multicellular organisms are genetically identical while the phenotypic expression that leads to these cells and tissues is very divergent. In other words, mechanisms need to be in place to select different fractions of the genome to be phenotypically expressed in e.g. liver cells and retinal cells, which requires meticulous gene regulation. Gene expression can be regulated at different stages, including RNA processing, translation, post translational modification of proteins, folding, signal transduction,… all of which contribute to the phenotypic outcome. However, one of the most fundamental regulations occurs at the transcriptional level where the actual gene is selected that codes for a specific protein. Epigenetics – roughly translated to “on top of” or “around” genetics – is the study of heritable changes of the phenotype resulting from reversible changes in gene function without altering the DNA sequence. It has such a profound impact on gene expression that it is stated by Goldberg et al. that “cellular differentiation may be considered an epigenetic phenomenon, largely governed by changes in the epigenetic landscape rather than alterations in genetic inheritance”(1). These changes can be caused by external factors, such as the metabolic state of the cell, or be part of normal development of the organism.
Workflow
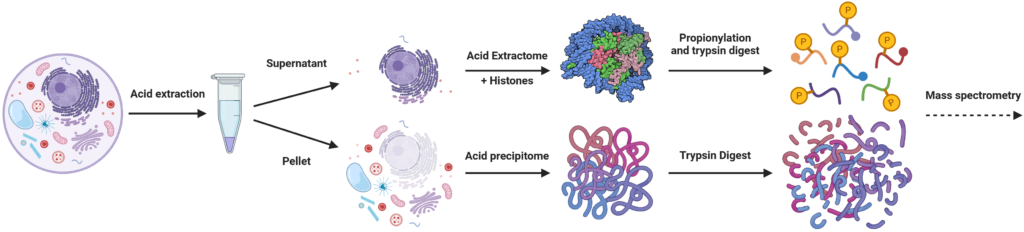
Our histone workflow allows to measure both the histone epigenetic and protein fraction on a single sample. This not only saves time and resources, but also enables the interpretation of histone epigenetic changes combined with altered protein expressions. To process the samples we start with an acid extraction of the histones and other basic proteins – the so-called acid extractome. This precipitates the rest of the proteome, which can be processed separately. This process is necessary not only to increase the coverage of the histone fraction, but also because histone proteins require a different sample preparation approach. This includes a lysine derivatization step to block these sites from digestion with trypsin, which is necessary to obtain peptides of sufficient length. Alternatively digestion can be done with an arginine specific enzyme (2). The precipitated protein fraction is processed with the regular suspension trapping protocol.
In total this workflow covers over 100 PTM sites and roughly 5000 quantifiable proteins.